Recently, I submitted my thesis project, which was a culmination of my Master’s course in Machine Learning at UCL. This is my second major project combining AI with medical imaging, and I had a great experience collaborating with my supervisor, Dr. Nikolas Pontikos, and everyone at Pontikos Lab.
Specifically, my project focused on Generative Adversarial Networks (GANs) for creating synthetic retinal images of inherited retinal diseases. Let me take you through this, starting with what inherited retinal diseases are.
What are Inherited Retinal Diseases?
Inherited retinal diseases, or IRDs, are a diverse group of genetic diseases known to cause visual impairment and blindness. Some people can have them early on from birth, while in others, the disease can develop progressively over time. IRDs affect around 1 in 3000 people in the UK, so they are super rare and can be debilitating because currently, the amount of treatment and interventions available is limited.
An example of an IRD is Stargardt’s Disease, which is relatively the most common of all rare diseases. People who have this disease present with a buildup of fat deposits on the center of the eye (also known as the macula), which is bad because this region is where most light focuses and our vision forms. How does this disease develop? Basically, people with Stargardt’s disease are known to have a mutation in a gene called ABCA4. This gene normally creates a protein that clears away a toxic molecule produced when the body manufactures specialized retinal cells (called photoreceptors). But when ABCA4 gets mutated, it “switches off,” and as a result, no protein will be created to get rid of the toxic molecule. This downstream leads to the buildup of toxic products, which combine with other molecules to form fat deposits.
Another example of an IRD is called vitelliform macular dystrophy or “Best” disease. Here, a gene called BEST1 is mutated. This gene makes a protein called Bestrophin-1, which nourishes the retinal epithelium, a layer of tissue that maintains the growth and development of the retina. Mutation of BEST1 is also known to cause abnormal fat buildup, damaging the photoreceptors and leading to poorer vision.
Like Stargardt’s disease and Best Disease, there are more than 200 identified IRDs. They can be categorized in many ways, like whether they affect people early on or are progressive, or whether they affect macular cells or some other cell type. The most common classification clinicians use is based on the genetic diagnosis, i.e., the mutated gene in the patient. But how does one find this out? Genetic screening is currently the #1 option available for patients, but there are many challenges. Firstly, identifying that the patient has a rare disease requires experienced clinical experts, who aren’t available in every clinic. Genetic screening is also accurate in only 40% of cases, as shown in this study. So clearly, there’s a lot of work to be done on this front, and we need to find better ways to improve diagnosis and care for IRD patients.
What my team is doing currently
As you may know, many deep learning applications have been developed which have very accurately diagnosed diseases from analyzing medical images. In our case, while IRDs are genetic by their pathogenesis, doctors have identified specific features in retinal images that visually suggest what the underlying mutation is.
The Moorfields Eye Hospital IRD dataset was recently compiled, containing roughly a million patient images and electronic records for 120 types of IRDs. An ongoing project is building a diagnostic model that uses deep learning to diagnose the underlying genetic disease, and some initial models have been trained already. But we found a problem: because some types of IRDs are rarer than others, the proportion of disease images of each gene in the dataset is imbalanced. This is not a good sign because typically, if you want a deep learning application to predict well on all disease classes, you’d like a relatively equal number of images of each class, but that’s not the case here. This is where GANs and I enter. In my project, SynthEye, I tried to implement a GAN that could generate synthetic images of the disease classes to augment our dataset and train better models in the future.
For an in-depth treatment of what GANs are, do check out my previous posts shared below!
But in short, GANs are unsupervised deep learning models that learn how to produce new image samples. The framework has two neural networks – a generator and a discriminator. The generator’s job is to take some random noise as input and transform it into a synthetic image. The discriminator’s job is to look at the generated image and classify whether it looks real or fake. Using the discriminator’s prediction of image realism as feedback, the generator tries to create more and more realistic samples with experience, such that the discriminator is fooled over time. It’s like I try to paint a picture of a dog, and my friend gives me feedback about whether my painting looks realistic or not. Over time, after my friend gives more and more feedback, I can create a picture so real that he will think it’s a dog photograph and not a painting!
Now you may be thinking, if an imbalanced dataset is not good for training a diagnostic model, how will it work with a GAN? Well, what makes a GAN, a generative model, different from a disease classifier, a discriminative model, is that the GANs objective is to understand properties of the different disease classes individually. In contrast, a classifier learns the bare minimum needed to distinguish between the classes. Putting it as an analogy, if you gave me a picture of a dog and a cat as asked me to classify them, all I’d have to look at is some key features like the size of the nose, whether the ears are pointy or not, etc. This is a discriminative model. But if you asked me to paint a realistic picture of a dog, I’m forced to focus on every small detail about the dog, not just the shape of the ears or nose, but everything from the amount of hair, the texture of the hair, the eye color, the tail, etc. This is a generative model. With such models, we can assume that despite the class imbalance, they will try to study each class separately; hence they would work well.
And with that, let me now take you through how I executed my project!
My Experiments
Within ophthalmology, there are different types of imaging that visualize the retina. The most common is fundus photography, where the optometrist shines a light in your eye and takes photos of the inside. This works well for a general assessment of the eye, but for more complex diseases, three other modalities are much better: fundus autofluorescence (FAF), infra-red (IR), and optical coherence tomography (OCT). I won’t go into details of how these imaging techniques work but at the core, what makes them unique is that they create images by harnessing reflectance properties of the retinal tissue, i.e., the chemical properties that arise when the tissues interact with light of different wavelengths. This means that if the disease has features like fat deposits, these interact differently than healthy tissue, so the image contrast is significant. Also, retinal diseases don’t just affect the topmost layer of the retina but even the layers underneath. Methods like OCT give this depth information as well.
As a starting point, my project just focused on generating synthetic FAF images. See an example of a FAF image below:
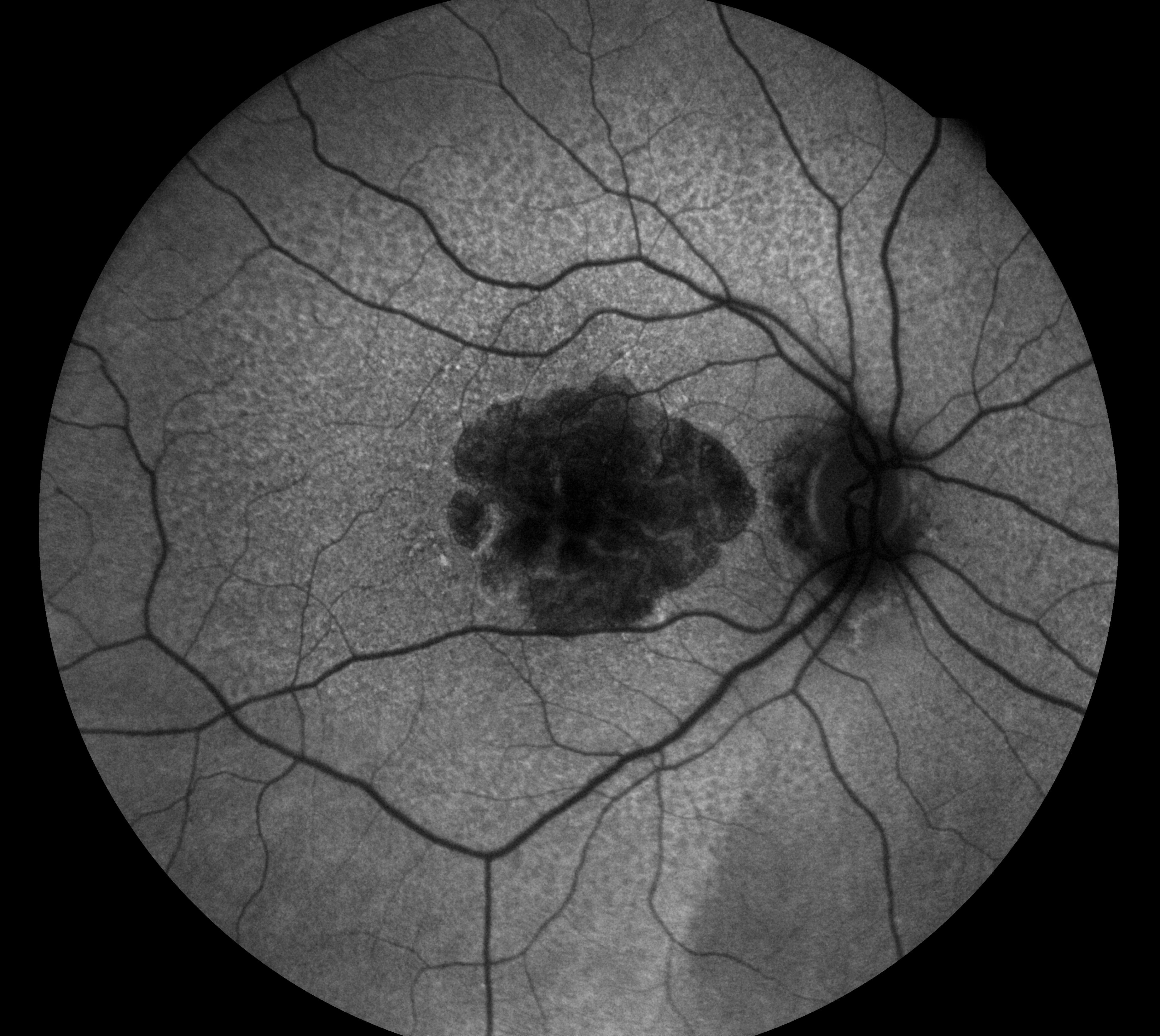
I used a dataset of around 15,000 FAF images associated with 36 IRD classes to develop my model. These images were from the same Moorfield’s Eye Hospital dataset as mentioned earlier.
Note that my goal was to not only generate photorealistic FAF images but I also had to generate the correct IRD gene. To do this, I slightly modified an existing state-of-the-art model called the multi-scale gradient GAN (MSGGAN). I’ll first discuss what this model is. If you’re not too familiar with deep learning, feel free to skip directly to the results!
Multi-scale gradient GANs
The MSGGAN was designed for generating high-resolution images. With the early GAN models like DCGAN and WGAN, one problem was that they struggled to generate images beyond 128×128 resolution and were unstable to train. To address this, the MSGGAN was one of the proposed methods to improve training. The model’s architecture is shown below, where the first half (the pink boxes) forms the generator and the second half (orange-green boxes) forms the discriminator.
What makes this model structure unique is the skip-connections between the generator and discriminator, shown as the black arrows. In the earlier GANs, the basic framework was that the noise would enter the generator, out comes the image, that image goes into the discriminator for predicting real/fake, and then to update the weights, the learning algorithm would backpropagate through the whole network. But this is different – here, you can see that the first box on the left (4 x 4 x c1) is projected into RGB space (using the red box under it), which is then fed to the discriminator end. This happens subsequently for the (8x8xc2) and (16x16xc3) and so on. What’s unique about this is that the model is training in a gradual manner where it first tries to master a 4×4 generation, then an 8×8 generation, then 16×16, all the way up to the highest resolution – and this is done simultaneously! The benefit of this type of framework is that the discriminator is now giving feedback (as gradients) at every “scale” or every resolution level, which stabilizes the training. Hence, it is known as “multi-scale gradients” GAN.
So we now know how the MSGGAN works. But this is just half the battle. I now had to adapt this architecture to my task of generating images of specific types of IRDs.
Making MSGGAN generate specific classes
Generating images “conditionally” or based on certain given information (like the classes) is an active area of research. For a start, I tried two standard methods: one-hot-encodings and embeddings.
- One-hot-encodings: A one-hot-encoding is a basic way to represent class information. The class is represented as an array of length N, where N = the total number of classes in the dataset. Each position in the array stores a bit (1/0), indicating which class is “hot” or active. So for example, let’s say I have 4 classes: [“cat”, “kitten”, “dog”, “houses”]. If I want to encode the “dog” class in the 3rd position of the list, the one-hot-encoding is [0, 0, 1, 0]. This vector can be attached to the GAN’s noise vector input and the discriminator’s image input as a way to force the model to generate that class. For more details, you can check out my earlier post on Conditional generation. I called this model CMSGGAN-1 (Conditional MSGGAN version 1) in my study.
Improving the GAN Part 2: Conditional and Controllable generation
2. Embeddings: One of the problems with one-hot-encodings, which I’m sure you can appreciate, is that as the number of classes gets large, the size of the encoding will also get large, and we are just storing vectors with a lot of 0’s (or sparse vectors), which is wasteful of computer memory. The value in the class vector also indicates the active class, which is not a very meaningful representation. Embeddings tackles this problem by instead using a semantic representation for each class. Specifically, embeddings are low-dimensional vectors where each value is some arbitrary attribute of the class. For example, for the same four classes as earlier, I could represent a 7-dimensional embedding as shown below. The row is the animal/object, while each entry describes some feature of the object like whether it’s a living thing, whether it has a gender, whether it’s plural etc.

Of course, these attributes are not the same as how a machine interprets them; instead, it will learn its own internal representation during training. I thought this was worth studying because embeddings have a fixed dimensionality (as the number of classes increases, the embedding vector is still the same size), and they will capture more semantic content of the classes rather than just a 1/0 in the one-hot encoding. Therefore, I implemented this as a second model variant called CMSGGAN-2 (Conditional MSGGAN version 2).
Results
Summarizing my methods, I trained two types of GANs, which I called CMSGGAN-1 and CMSGGAN-2. The only difference between both models is how I forced it to generate the image of a specific IRD class. Overall, I found in my experiments that CMSGGAN-1 was the best-performing model. Below, I show some of the examples of generated images from the model. Each row shows an IRD gene that’s mutated, and each column is a different image generated by a different noise vector input to the GAN. The images look small, but they are 256×256 resolution.

I’m sure you can notice how the features of the image look similar across the columns? Well, that is because each column has the same noise vector but a different class encoding. As a result, the generated images have similar structures like the blood vessels and the background intensities. But some subtle details make them look different, and these are associated with the disease information that comes in the class encoding. For example, BEST1 has a white yolk-like appearance in the center of the retina, which is characteristic of Best disease, and ABCA4 has white spots representing the fat deposits.
The above images were still photorealistic, though – I found some images that just looked really trippy and could clearly be called fake. For example, see below how the images have two sets of blood vessels connecting? This is biologically impossible! There are also some images where blood vessels come from nowhere, which is also nonsensical.
 |
 |
 |
 |
Setting aside these anomalies, for now, if we just consider a generated dataset as a whole, there’s one crucial question: how can we be sure the images accurately represent the correct diseases and features as in the real dataset? Especially if we are going to use it for augmenting our existing imbalanced IRD dataset, we want to be absolutely sure that all the generated data is correct, right? This is one of the major challenges in generative modeling research, and people are actively developing better ways to evaluate results. The most basic strategy, of course, is by showing it to an expert who can visually inspect and comment on the appearance. Unfortunately, I couldn’t get a visual evaluation before submitting my thesis, but I hope to do this now since it is the gold standard.
The next best option was using existing metrics of image quality. For my first metric, I wanted to make sure that the generated images were not memorized from the training set. Memorization is bad because it indicates that the model hasn’t really learned anything. To measure this, I compared every one of my generated images with the actual images from the training set using Pearson’s correlation coefficient. For every real/generated image pair, if the correlation is exactly +1, the images are the same, and if lesser than 1, then there is some difference. This is shown below:

For my generated images, I found that most of the correlation values were around 0.6-0.8, which is close to +1 but not exactly +1. This is a good sign because it means that the images were diverse to some extent. Also, diverse images are not tied to a specific patient, so that means we can freely generate new images without having to worry about issues like data confidentiality.
Another metric I tried was the Frechet Inception Distance (FID), which compares the distribution of pixel intensities of the generated images with real IRD images. Generally, a lower FID value means better performance of the GAN. Comparing CMSGGAN-1 and CMSGGAN-2, I found that the CMSGGAN-1 model had a lower FID of 50 while CMSGGAN-2 had a value of 60, which meant the former’s images were much better. However, I still found this metric hard to interpret because there was no baseline on what is considered a good FID. Aside from this, I tried some other metrics that you can find in my report below if interested.
Finally, I wanted to try a fun experiment typically done in the popular GAN papers – exploring the latent space. The idea is that if you modify the noise vector or the class vector in a specific way, you can change certain features in the image. For example, by modifying the noise, you can morph a starting face into other different faces, like below.

While this is just a fun experiment with face-generating GANs, this could be super useful in the medical field. For example, if you want to understand the differences between disease features, we could convert one disease image into another. This can be a great educational tool for doctors or medical students! Or suppose you want to represent how someone’s brain looks if it had different stages of Alzheimer’s disease? You could take a starting brain scan, transform it into a latent space vector, and then modify it to create the same brain in different disease stages. This would be amazing for disease progression modeling. So with that, I finally share these two cool gifs of retinal images below. The first image shows how Stargardt’s disease morphs into Best disease. The second image shows how one example of Stargardts disease morphs into another example – in this case, it’s more of a right eye to left eye transition. I found this quite remarkable because you can see how there’s a sharp transition from left to right – it means that the GAN has learned that there can’t be a right + left eye – it’s either right or left.


Looking back…
This study is just the beginning of my exploration of GANs. There is still a lot of work to do in terms of evaluating the images and also training the models to generate at even higher resolution. I also hope to combine this data generator with the diagnostic model and retrain it to improve its performance.
Overall, this project has been a great learning experience for me. Reflecting on the past three months, I would say that the process of studying these models and reading the literature was the most enriching period. The Coursera GAN Specialization was extremely helpful in simplifying complex ideas for me, and I would recommend it to anyone who wishes to study GANs in-depth. Funnily, almost two years back, I remember writing an article about GANs in my old blog but rereading it, I realized I probably didn’t understand the beauty of how these models learn. My MSc Machine Learning course really changed me by honing my mathematical and computational skills, which allowed me to appreciate better how stuff works under the hood. I’ve now realized that this is an area I want to study further in my future research journey.
This project has also helped me appreciate better how one can effectively use AI in medicine. Looking from a bird’s eye view at the research landscape, one thing I’ve noticed is that when it comes to more common diseases like diabetes, cancer, or cardiovascular pathology, there has already been so many years of learning and expertise developed in the medical community that clinicians have been a bit hesitant to bring in AI innovations. But learning more about rare diseases like IRDs, for which the knowledge is quite limited, I think AI could be a valuable aid for clinicians to provide better care to patients, and I think this is a space where AI will take off well.
Specifically, I believe GANs and generative modeling, as a field, will be revolutionary in how we process and analyze medical data. As the renowned physicist Richard Feynman said, “What I cannot create, I do not understand.” Generative models have the ability to create, hence understand data probably better than we do, and I think combining some of this machine understanding with our human understanding is what will make medicine better in the future.
If you’re interested to read my paper, do check it out below! Also, code is shared in this GitHub repo.