In 2018, my participation in the COSMOS summer program “Mathematical Modeling in Molecular Biology” at UC Davis first introduced me to the field of interdisciplinary. During the course of five weeks, I was immersed in lectures, tutorials, and wet lab sessions, all seeking to integrate Knot theory with molecular biology. In class, I was engrossed in the power of KnotPlot, a software that can be deployed to analyze the topological features of biomolecules. To obtain a wholistic experience of interdisciplinary research, my peers and I coded a program in KnotPlot to attain the mathematical knot type of a DNA sequence extracted from bacteriophage P4, then validated the computational result in a lab by agarose gel electrophoresis. Both the computational and experimental results showed trefoil being the major knot type. We explained this phenomenon by the confinement of P4 DNA within the phage capsids. From this experience, I realized that programming builds the bridge between “ATGCs” and “XYZs;” my enthusiasm for interdisciplinary research flourished and my mindset as a future researcher was cultivated.
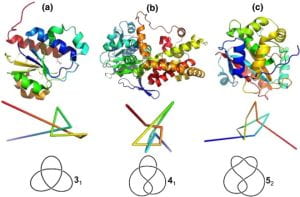
Following this program, I continued developing this newfound academic interest by conducting a research on protein topology under the supervision of two UC Davis professors – Prof. Javier Arsuaga, Prof. Mariel Vazquez, and Dr. Rob Scharein – developer of a topological software, KnotPlot. My project analyzed the topological features of Leucine Rich Repeat Kinase 2 (LRRK2) mutants, which are associated with an increased risk of Parkinson’s Disease. A Java program was developed for protein backbone extraction and KnotPlot deployed for topological analysis which indicated that the unknotted wild type of protein became a trefoil knot in the case of some pathogenic mutations. This intriguing result propelled me to uncover the correlation between the change in topological properties and the malfunction of the mutated protein. Seeking an opportunity to discuss my project, I delivered a poster presentation titled “Topological Analysis of Protein Folding of LRRK2 Mutants in Parkinson’s Disease” at the 2019 World Life Science Conference in Beijing and was fascinated by the topics presented by mature scientists and the advice they offered me.
Currently, my focus is on applying computational biology and bioinformatics to study the functional and structural significance of knotted proteins. Some ongoing projects are: (1) creating catalogues of knotted proteins by knot type, protein function, and disease association; (2) developing a GNN to classify protein knot type using topological features from persistent homology; (3) studying the relationship between the topological features and functions of knotted proteins. Overall, the aims are to better understand why certain proteins are knotted and how the folding of knotted proteins can offer insights into the folding of all proteins.

